In every computing device, there lies a small silicon chip used to help transmit current to different components. The major rule of computing production, Moore’s Law, states that every two years, the number of transistors on a silicon chip will double. This doubling is pivotal, as an increased transistor count enhances computational speed, allowing computers to process tasks concurrently.
In order to reduce production costs, businesses attempt to shrink the silicon chips while also meeting the principles of Moore’s law. The method of shrinking chips and overflowing them with transistors has worked for a while, but now companies face a roadblock: they can’t shrink silicon chips and keep up with Moore’s law. So what now?
Discovered by Professor Leonard Adleman of USC in 1994, DNA computing serves as a plausible solution to our problems as it has the potential to create a near-zero carbon footprint, be cost effective, improve security and last a long time with very little maintenance.
DNA computing uses chemicals to make synthetic DNA and DNAzymes, enzymes such as DNA ligase that assist in the replication of DNA, in place of electronics, to perform data storage and computations. In this process, data undergoes transformation into DNA’s four nitrogenous bases—adenine (A), thymine (T), cytosine (C), and guanine (G)— and is housed in a small test tube.
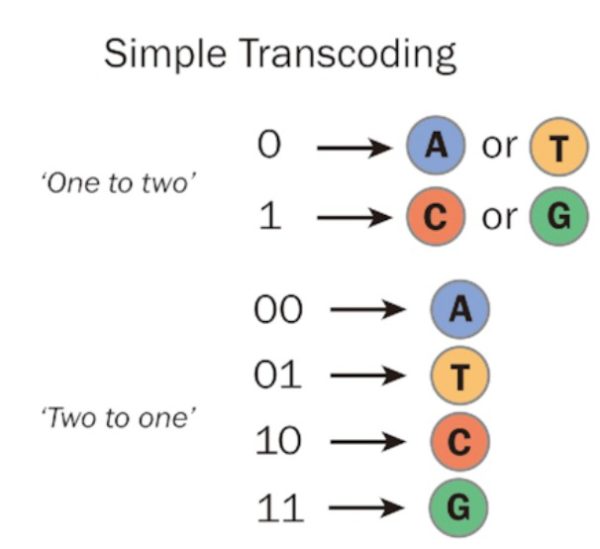
To convert binary data into DNA, the binary information is initially partitioned into pairs of bits. In this process, there are four distinct ways to create unique combinations of 1s and 0s, each pair having a length of two. These four pairs are encoded into the DNA bases using a key:
DNA’s unique feature called Watson-Crick pairing, where base A bonds to only base T, base C bonds only to base G, and vice versa, is essential when it comes to processing the newly synthesized DNA strands. In DNA computing, processing usually involves unzipping the double helix into two single strands then joining the strands together.
Processes such as amplification, gel electrophoresis, and the Biotin-Streptavidin system are used in the retrieval of the output. Amplification is where single strands of the DNA are replicated many times to make them abundant in the solution. In gel electrophoresis, electric current is used to move charged molecules through a gel matrix; smaller molecules usually move further away from the starting point than larger molecules do.
Through using this process, DNA with the desired length is separated from the original solution. The Biotin-Streptavidin system uses biotin, also known as vitamin H and in this case used to attach to molecules of interest, and streptavidin, a protein from bacteria that has a very strong affinity for biotin.
Using these two molecules and their affinity for each other, the Biotin-Streptavidin system uses labeled biotin molecules to attach to specific strands of DNA, and has the streptavidin bind to these biotin molecules; this allows the most closely related DNA strands to be collected. Once the result is obtained, the sequence of As, Gs, Ts, and Cs is decoded back to binary and can be used for display purposes.
DNA computing has been said to one day overtake the silicon chip, but currently it still has a long way to go in terms of speed and potential affordability. In terms of speed, DNA computing can take over 10 hours, whereas the silicon chips we currently use run in nanoseconds. This is because in DNA computing, all possible solutions to the question are developed and sorted through, whereas in a silicon chip only the correct answer is produced.
In terms of cost, DNA computing costs $7,000 to synthesize the 2 megabytes of data in the files, and another $2,000 to read it, whereas the average computer can store and read 8 gigabytes of data for around $1,000.
DNA computing is still a developing technique that may become the future of computing. Currently, a few DNA computers have been used in labs for problems that involve complex combinatorial optimization or pattern recognition.
Additionally, researchers at universities such as MIT, the University of Washington, and more have found that image, digital, and textual data can be stored in DNA. With companies such as Microsoft, Google, Meta, and so many more investing millions of dollars into DNA computing, you could someday be reading this using a DNA computer.


